| IMIM |
| UPF |
| CRG |
| GRIB |
| SOFTWARE |
| gff2aplot |
| Genome BioInformatics Research Lab |
| Help | News | People | Research Software Publications | Links |
| Resources & Datasets | Gene Predictions | Seminars & Courses |
|
gff2aplot |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Contents
gff2aplot
|
|
|
Abril, J.F., Guigó, R. and Wiehe, T. " gff2aplot: Plotting sequence comparisons." Bioinformatics, 19(18):2477-2479 (2003). [Bioinformatics Abstract] [PubMed Abstract] URL: https://genome.crg.es/software/gfftools/GFF2APLOT.html ...Thanks in advance for your collaboration. |
| EXAMPLES |
|
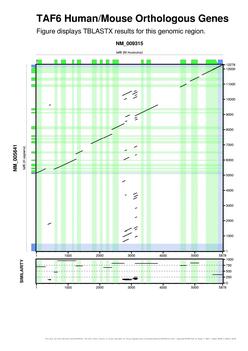 | Following this link you can get some snapshots of gff2aplot output. |
You can find several examples of the flexibility of gff2aplot in those of the following references which include the figure numbers where the program was used (those published before 2004 were made using the old GNUawk/Bash implementation).
Do you think we have forgotten any citation ? Do not hesitate to send an email with the corresponding citation to authors and we will include it here...
| HOWTOs |
|
In this section you can find usefull tutorials on how to use gff2aplot. It will be regularly updated with new documents. Some knwoledge about unix commad-line is required, more specifically about bash shell.
Most of the snapshot figures from those HTML pages are in PNG format. Our apologies if your browser is not able to handle PNGs. Anyway, you will find a link to a PostScript and/or PDF file for the main figures.
parseblast output when applying to a WU-BLAST file. There are three basic aligment formats that can be generated from a blast file by parseblast.pl, but all three must produce the same plots by gff2aplot. We will also see the raw output from gff2aplot and how to customize it a little bit. ali2gff, a filter written in C, to convert its output into GFF. Once we have generated the whole genomic region figure, we will zoom into a single gene annotation. This will illustrate how you can expand a given area from your pictures, to highlight single features on multigenic annotation plots for instance. gff2aplot allows to emulate color blending to enhance the visualization of those projections. gff2aplot layers: gff2aplot is put into one of the pre-defined layers. It is obvious that it is important to know which layers we have and what can be shown on each of them. Another interesting feature that can be learnt from this tutorial, is the way gff2aplot can handle customizing parameters from command-line and customization files. You are welcome to provide more examples on how did you use gff2aplot in your projects, by sending your report files or a link to your own html report/howto. Your experience will be valuable for other users, mostly for newer ones. Send an email to authors, we will try to include here your contribution as soon as possible.
| NEWS |
|
| 12 Dec 2003 | v2.0 |
Applications Note for gff2aplot has been published in Bioinformatics (it was submited on March 4, 2003; revised on June 11, 2003 and finally accepted on June 20, 2003).
See program description section for a complete reference. |
||
| 02 Jun 2003 | v2.0 |
Finally, gff2aplot has been fully re-implemented in Perl. Parsing process, record sorting and output throughput is much more faster. We also expect that the new program will be more portable than the previous versions. Customization process is now more flexible, including gff2ps-like customization files (regular expressions, modularity of settings and multiple files allowed), ability to set all the variables via command-line switches. Output PostScript code has also been improved, making the plots more compact and less prone to interpreter errors. Old GNUawk/Bash version is no longer maintained although it is still available. |
||
| 26 Jan 2000 | v1.9 |
After several bug fixes and versions not published, here is the new version for gff2aplot. This version can work with "partial" custom files (without having to define all the variables) and can also read alignments in standard GFF format. |
||
| 24 Apr 1999 | v1.3 | Defaults are defined within the main gawk script too. Thus custom file is not mandatory now. | ||
| 22 Mar 1999 | v1.2 | Zoom option was defined. Data vectors can be passed to plot functions on the third panel. | ||
| 02 Mar 1999 | v1.0 |
First running version for gff2aplot. Annotation on axes is defined in GFF-format. Alignment definition is written in pseudo GFF (APLOT format). |
| DOWNLOADING |
|
Download last version of gff2aplot (v2.0) from our ftp server. The last version of the "gff2aplot User's Manual" is also available via ftp altough it is included in the program's tarball. This manual is not finished yet, you still can learn how to use gff2aplot from the examples described at the tutorials section. From version 2.0 we are only developing the perl implementation. The old GNUawk/Bash is no longer maintained but you can still get it from this link to gff2aplot version 1.9.
You have downloaded a gzipped tarball containing the perl version gff2aplot, all the supplementary scripts (i.e. WU-BLAST/NCBI-BLAST/SIM/BLAT filters), a README text file and few examples of what can be done with this tool. You can extract them with:
gunzip -c gff2aplot-vX_xx.tar.gz | tar xvf -
On a Linux box you can try with:
tar zxvf gff2aplot-vX_xx.tar.gz
A gff2aplot-vX_xx directory will be created, move into that directory and take a look to the README and INSTALL files. Once you have read them, just type:
make
and then:
make install
By default, the last command will move all the scripts to /usr/local/bin but if you want to place all the exec files into another directory, just define the new installation path as follows:
make INSTALLDIR=/your/path/bin install
Another way of doing that is modifying that variable on the Makefile accordingly to your needs.
Sorry but there is not a "make test" yet...
At this point you are ready for running gff2aplot.pl and related code, see examples on how to use it from the tutorials section.
| REPORTING BUGS |
|
If you find any bug or something is not plotted properly, you can send a bug report. To easily find what's wrong, you should attach to that e-mail a tarball containing the custom file you were using when the bug ocurred, an example of your input GFF files, the PostScript file generated and a report file that you can get with the "-V" command-line option (type "gff2aplot -h" for further info on that verbose mode switch). We will try to answer as soon as possible.
| FEATURE LIST |
|
The following menu lists many features of gff2aplot:
| WISH LIST |
|
Although we have implemented many features, there are few ideas not ready yet for the current version of gff2aplot.
Here is a short list:
We are open to any helpful suggestion for improving our programs. Do not hesitate to get in touch with us.
| AUTHORS |
|
| Disclaimer | webmaster |